Difference between revisions of "BrainStreamExampleSentences"
Wiki-admin (talk | contribs) |
Wiki-admin (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 6: | Line 6: | ||
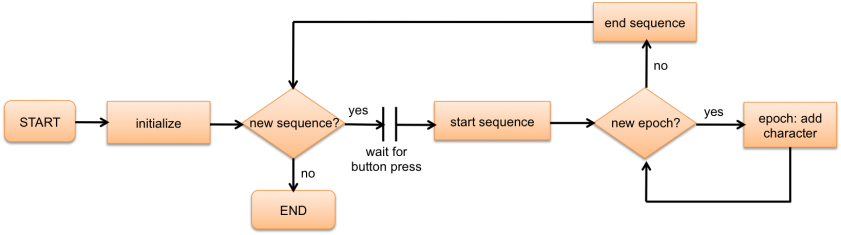
This is an example of a very simple 'experiment' in which letters will appear on the screen one by one to form sentences. In this experiment, adding a new character to a string is called an ''epoch''. A series of epochs that results in a complete sentence is called a ''sequence''. Subjects can press a button when they are ready for the next sentence (sequence). A flowchart of the experiment is shown below: | This is an example of a very simple 'experiment' in which letters will appear on the screen one by one to form sentences. In this experiment, adding a new character to a string is called an ''epoch''. A series of epochs that results in a complete sentence is called a ''sequence''. Subjects can press a button when they are ready for the next sentence (sequence). A flowchart of the experiment is shown below: | ||
| − | [[File: | + | [[File:DocsSectionsExampleSentences_Flowchart.png|841px|235x]] |
''Figure 1: Flowchart of the Sentences example experiment'' | ''Figure 1: Flowchart of the Sentences example experiment'' | ||
| Line 279: | Line 279: | ||
=== Creating the Dictionary table === | === Creating the Dictionary table === | ||
| − | The [ | + | The [[BrainStreamBuildingExperiments#SecDict|Dictionary table]] specifies a marker number for each marker that is used in this experiment. The Dictionary table looks like this: |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 300: | Line 300: | ||
|} | |} | ||
| − | In this experiment, the fourth column ''datasource'' is not absolutely necessary. As only one data source (simulated EEG data, see [ | + | In this experiment, the fourth column ''datasource'' is not absolutely necessary. As only one data source (simulated EEG data, see [[BrainStreamExampleSentences#BrainStream|blocksettings]) is used, BrainStream will automatically apply all marker settings to this data source. |
== Testing the experiment == | == Testing the experiment == | ||
| Line 306: | Line 306: | ||
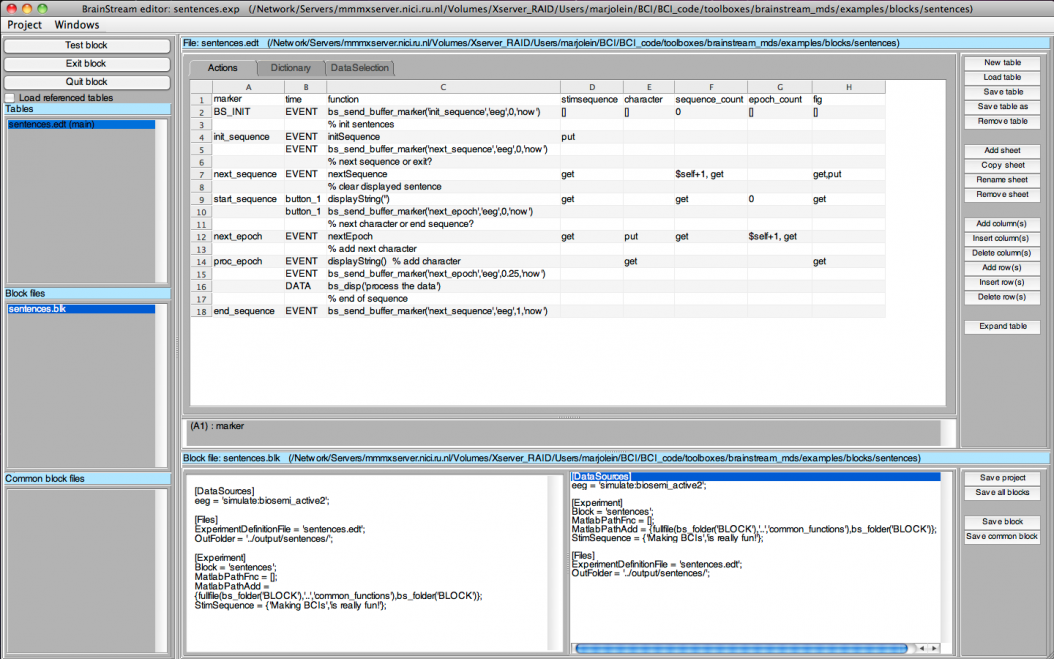
The easiest way to create all files that define this experiment is via the [.DocsSectionsBrainStreamEditor BrainStream editor]. You can open the editor by typing <tt>bs_editor</tt> in the Matlab command window (make sure that <tt>brainstream_mds/core</tt> is your current working directory). An experiment file (.exp) has already been created for this example experiment and is located in <tt>/brainstream_mds/examples/blocks/sentences/sentences.exp</tt>. If you open this file in the BrainStream editor, the experiment looks like this: | The easiest way to create all files that define this experiment is via the [.DocsSectionsBrainStreamEditor BrainStream editor]. You can open the editor by typing <tt>bs_editor</tt> in the Matlab command window (make sure that <tt>brainstream_mds/core</tt> is your current working directory). An experiment file (.exp) has already been created for this example experiment and is located in <tt>/brainstream_mds/examples/blocks/sentences/sentences.exp</tt>. If you open this file in the BrainStream editor, the experiment looks like this: | ||
| − | [[File: | + | [[File:DocsSectionsExampleSentences_BS_editor.png|1054px|658x]] |
''Figure 2: The Sentences Experiment in the BrainStream Editor'' | ''Figure 2: The Sentences Experiment in the BrainStream Editor'' | ||
Latest revision as of 13:50, 26 September 2018
Example 1: Sentences
Experiment description
This is an example of a very simple 'experiment' in which letters will appear on the screen one by one to form sentences. In this experiment, adding a new character to a string is called an epoch. A series of epochs that results in a complete sentence is called a sequence. Subjects can press a button when they are ready for the next sentence (sequence). A flowchart of the experiment is shown below:
Figure 1: Flowchart of the Sentences example experiment
Building the experiment
As described in the Building Experiments section, a number of files are needed to define an experiment. In the following, we will show how to create each of these files. If you want to have a look at the final result before reading this tutorial, the experiment file (.exp) for this example can be found in /brainstream_mds/examples/blocks/sentences/sentences.exp.
Creating the block file
This experiment consists of only one block (sentences.blk). A number of settings are specified in the block file:
[DataSources]
eeg = 'simulate:biosemi_active2';
[Files]
ExperimentDefinitionFile = 'sentences.edt';
OutFolder = '../output/sentences/';
[Experiment]
Block = 'sentences';
MatlabPathFnc|Matlab Path Fnc = [];
MatlabPathAdd|Matlab Path Add = {fullfile(bs_folder('BLOCK'),'..','common_functions'),bs_folder('BLOCK')};
You can look up the meaning of the topic and key combinations here.
Note that for the keys that specify folders, such as OutFolder and MatlabPathAdd, relative path names are specified. For more information about relative path names, click here.
In addition to the standard topics and keys, we can specify our own settings in the block file. In this example, it would be useful to specify in the block file which sentences we want to show. We could for example define a key StimSequence under topic Experiment:
[Experiment]
StimSequence = {'Making BCIs','is really fun!'};
The information specified in the block file can be retrieved during the experiment using the bs_get_blockvalue function. In the next section we will show exactly how this works. The advantage of specifying the sentences in the block file is that we can use the same Actions table for displaying any sentence.
Creating the Actions Table
The most striking characteristic of this experiment is the need for decision making throughout the experiment. From the flowchart in figure 1, it can be seen that two types of decisions must be made:
- before each sequence, decide whether to start a new sequence or end the experiment
- before each epoch, decide whether to start a new epoch or end the sequence
We will write user defined functions for making these decisions.
In order to be able to make each decision, we also need to:
- specify how many sequences need to be shown
- keep track of how many sequences we have shown so far
- specify how many epochs are in each sequence
- within each sequence, keep track of how many epochs we have shown so far
We will make use of user defined variables for these tasks.
The Actions table has three standard columns marker, time and function. In addition, we will add three columns for our user defined variables. The first user variable, stimsequence, will contain the sequences that we want to show. The second user variable, sequence_count, will keep track of the number of sequences that have been shown already. The third user variable, epoch_count, will keep track how many epochs of a sequence have been shown already. These are the columns of the Actions table:
| marker | time | function | stimsequence | sequence_count | epoch_count |
We will initialize these variables at the beginning of the experiment. It is usually good to define the initialization actions at the BS_INIT marker, as this is always the first marker to arrive within an experiment. At the beginning of the experiment, the sequence counter will be set to 0. The epoch counter is initialized as empty and will be set to 0 at the beginning of each new sequence. The variable stimsequence must contain the sentences that will be shown. We will initialize it as empty and retrieve its content from the block file in the next step.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | [] | 0 | [] |
Information specified in the block file can be retrieved during the experiment using the bs_get_blockvalue function. At the BS_INIT marker, we will insert the marker init_sequence. As this marker does not need to be accurately synchronized with the data, we insert this marker via the FieldTrip buffer. When the init_sequence marker arrives, we will execute a user defined function initSequence to retrieve the content of stimsequence from the block file and place it in the global variables.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') |
[] | 0 | [] |
| init_sequence |
EVENT |
initSequence |
put |
This is the initSequence function:
function event = initSequence(event)
event.stimsequence = bs_get_blockvalue('Experiment','StimSequence',{'Making BCIs','is fun!'});
The bs_get_blockvalue function will get the content specified under topic Experiment and key StimSequence and will use the default sentences 'Making BCIs' and 'is fun!' if no specification is found in the block file. The value that is retrieved from the block file will be placed in the field event.stimsequence. Because a put statement is specified for this variable in the Actions table, this new content will be updated to the global variables from where it can be accessed later in the experiment .
Now we can proceed to the next step in the flowchart, the point where we need to decide whether to start a new sequence. Using the bs_send_buffer_marker function, we will insert a marker next_sequence to begin this next stage of the experiment. When this marker comes in, first the sequence counter must be increased by 1:
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | $self+1 |
Subsequently, we need to write a function that uses the content of stimsequence and the current value of sequence_count to decide whether to start a new sequence. We will call this function nextSequence. Because the function needs the content of two of our variables as input, we must specify get statements for these variables.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get |
This is function nextSequence:
function event = nextSequence(event)
if event.sequence_count <= numel(event.stimsequence) % if sequence_count <= number of sequences
event = displayIt(event,'string','Press button_1 (<ctrl>-b) to start sequence',[1 0 0]); % display instruction to press button
event = bs_send_buffer_marker(event,'start_sequence','eeg',0,'now'); % start this sequence
else % all sequences processed
event = bs_send_buffer_marker(event,'BS_EXIT','eeg',0,'now'); % exit block (insert BS_EXIT marker)
end
The first input argument of a user defined function is always event. The content of variables for which a get statement is specified, is copied into fields of the event structure. Thus, event.stimsequence contains the content of stimsequence and event.sequence_count contains the content of sequence_count.
If the outcome of the decision is that another sequence should be displayed, the marker start_sequence will be inserted. At the start of a new sequence, the variable epoch_count must be set to 0. Then BrainStream must wait for a button press from the participant, which is achieved by specifying timepoint 'button_1' for this event. The displaying of the characters is handled by the function displayString. This function will not be discussed in detail, but it is important to know that it requires the content of the variables stimsequence and sequence_count. Therefore, get statements must be specified for these variables. Now we have reached the next decision point in the experiment flowchart, where we need to decide whether to start a new epoch or end the sequence. We will insert marker next_epoch to trigger this decision making step.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get | |
| start_sequence | button_1 | displayString() | get | get | 0 |
| bs_send_buffer_marker('next_epoch','eeg',0,'now') |
When the next_epoch marker comes in, first the epoch counter must be increased by 1. Subsequently, we need to write a function that uses the content of stimsequence and the current values of sequence_count and epoch_count to decide whether to start a new epoch. We will call this function nextEpoch. Because the function needs the content of all our variables as input, we must specify get statements for these variables. The Actions table now looks like this:
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get | |
| start_sequence | button_1 | displayString() | get | get | 0 |
| bs_send_buffer_marker('next_epoch','eeg',0,'now') | |||||
| next_epoch | EVENT | nextEpoch | get | get | $self+1, get |
This is function nextEpoch:
function event = nextEpoch(event)
if event.epoch_count <= numel(event.stimsequence{event.sequence_count}) % if epoch_count <= number of epochs in this sentence
event.character = event.stimsequence{event.sequence_count}(event.epoch_count); % this line is required for displayString function - ignore
event = bs_send_hardware_marker(event,'proc_epoch','eeg'); % insert marker to process this epoch
else % all characters processed
event = bs_send_buffer_marker(event,'end_sequence','eeg',0,'now'); % end the sequence
end
If the outcome of the decision is that another character should be displayed, the marker proc_epoch will be inserted. As we want to use this marker for data selection (see [[BrainStreamExampleSentences#DataSelection|next section]), accurate synchronization between marker and data is required. Therefore, we will insert this marker [[BrainStreamMarkerHandling#HardwareMarker|via the hardware]. When this marker arrives, the displayString function will be used to show the current character. After this has been done, we insert the next_epoch marker and thereby return to the question whether to start another epoch. The next_epoch marker is inserted 0.25 seconds after the bs_send_buffer_marker function is called. As a result, there is a pause of approximately 0.25 seconds between the appearance of each character on the screen.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get | |
| start_sequence | button_1 | displayString() | get | get | 0 |
| bs_send_buffer_marker('next_epoch','eeg',0,'now') | |||||
| next_epoch | EVENT | nextEpoch | get | get | $self+1, get |
| proc_epoch | EVENT | displayString() | |||
| bs_send_buffer_marker('next_epoch','eeg',0.25,'now') |
When all epochs of a sequence have been processed, the nextEpoch function will insert the end_sequence marker. When this marker arrives, we immediately insert the next_sequence marker, thereby returning to the question whether to start another sequence. The next_sequence marker is inserted 1 second after the bs_send_buffer_marker function is called. As a result, there is a pause of approximately 1 second between each sequence. The Actions table now contains all the necessary steps of the experiment.
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get | |
| start_sequence | button_1 | displayString() | get | get | 0 |
| bs_send_buffer_marker('next_epoch','eeg',0,'now') | |||||
| next_epoch | EVENT | nextEpoch | get | get | $self+1, get |
| proc_epoch | EVENT | displayString() | |||
| bs_send_buffer_marker('next_epoch','eeg',0.25,'now') | |||||
| end_sequence | EVENT | bs_send_buffer_marker('next_sequence','eeg',1,'now') |
Specifying Data Selection
For BCI applications, you will often want to collect data associated with stimuli. In this example, you might want to collect one second of EEG data every time a new character appears on the screen. In your Actions table, you must specify at which marker you want to collect data. In a DataSelection table, you must specify exactly how much data you want to select and from which data source.
New epochs are started by the proc_epoch marker. We will therefore use this marker to trigger data selection. For now, we will only print the message 'Process the data' in the Matlab command window every time data is collected. However, instead of showing a simple message you could also write functions to actually process the data. This is the new Actions table:
| marker | time | function | stimsequence | sequence_count | epoch_count |
| BS_INIT | EVENT | bs_send_buffer_marker('init_sequence','eeg',0,'now') | [] | 0 | [] |
| init_sequence | EVENT | initSequence | put | ||
| bs_send_buffer_marker('next_sequence','eeg',0,'now') | |||||
| next_sequence | EVENT | nextSequence | get | $self+1, get | |
| start_sequence | button_1 | displayString() | get | get | 0 |
| bs_send_buffer_marker('next_epoch','eeg',0,'now') | |||||
| next_epoch | EVENT | nextEpoch | get | get | $self+1, get |
| proc_epoch | EVENT | displayString() | |||
| bs_send_buffer_marker('next_epoch','eeg',0.25,'now') | |||||
| DATA |
bs_disp('Process the data') | ||||
| end_sequence | EVENT | bs_send_buffer_marker('next_sequence','eeg',1,'now') |
We also need a DataSelection table to specify the exact time window of data selection. If we want to record one second of data after the onset of each epoch, our DataSelection table looks like this:
| marker | begintime | endtime | datasource |
| proc_epoch | 0 | 1 | eeg |
In this experiment, the fourth column datasource is not absolutely necessary. As only one data source (simulated EEG data, see [[BrainStreamExampleSentences#BrainStream|blocksettings]) is used, BrainStream will automatically apply all data selection settings to this data source.
Creating the Dictionary table
The Dictionary table specifies a marker number for each marker that is used in this experiment. The Dictionary table looks like this:
| marker | type | value | datasource |
| init_sequence | stimulus | 1 | eeg |
| next_sequence | stimulus | 2 | eeg |
| start_sequence | stimulus | 3 | eeg |
| next_epoch | stimulus | 4 | eeg |
| proc_epoch | stimulus | 5 | eeg |
| end_sequence | stimulus | 6 | eeg |
| button_1 | stimulus | 7 | eeg |
In this experiment, the fourth column datasource is not absolutely necessary. As only one data source (simulated EEG data, see [[BrainStreamExampleSentences#BrainStream|blocksettings]) is used, BrainStream will automatically apply all marker settings to this data source.
Testing the experiment
The easiest way to create all files that define this experiment is via the [.DocsSectionsBrainStreamEditor BrainStream editor]. You can open the editor by typing bs_editor in the Matlab command window (make sure that brainstream_mds/core is your current working directory). An experiment file (.exp) has already been created for this example experiment and is located in /brainstream_mds/examples/blocks/sentences/sentences.exp. If you open this file in the BrainStream editor, the experiment looks like this:
Figure 2: The Sentences Experiment in the BrainStream Editor
You will see that this Actions table has two additional columns: character and fig. These two variables are required for the displayString function and will not be discussed here. You can click the 'Test block' button to see how the experiment looks. In addition, try changing the StimSequence blocksetting and see how easy it is to show different sentences.