BrainStreamInstallation
Installation Guide
Start-up checklist
Ensure availability of a Matlab license
BrainStream requires [Matlab] version 2008a or higher. The availability of this license depends on your institution's licensing agreements with MathWorks. BrainStream also requires Matlab's Signal Processing Toolbox.
Ensure access to BrainStream code and required toolboxes
The BrainStream package can be downloaded here (only accessible by members) or via alternative checkout from another repository (internal people only).
BrainStream requires some additional software, which is standardly included with the distribution of BrainStream. This is an overview of the additional software:
FieldTrip toolbox
Java library for reading/writing Excel, jxl.jar GNU Library or Lesser General Public License (LGPL)
MSocket: Copyright (c)* 2006 MIT Lincoln Laboratory, Steven Michael (smichael@ll.mit.edu)
fparser.m: Copyright (c)* 2009, urs (us) schwarz (us@neurol.unizh.ch)
uipickfiles.m: Copyright (c)* 2007, Douglas M. Schwarz
inifile.m: Copyright (c)* 2003-2005 Primoz Cermelj, Slovenia (primoz.cermelj@email.si)
findjobj.m: Copyright (c)* 2014, Yair Altman
GUILayout: Copyright ©* 2014, Ben Tordoff
jsonlab: Copyright(c)* 2011-2014 Qianqian Fang
(c)* All rights reserved, BSD license (see Matlab for explanation or read the BSD.txt file in third_parties subfolder in BrainStream's core folder)
Add BrainStream's core folder to the Matlab path
The BrainStream core folder must be on your Matlab path. You can either add this folder to the Matlab path as described here or make sure that the BrainStream core folder is your current working directory.
During runtime, BrainStream itself will add all folders required for BrainStream processing to the path. If you need any additional folders on your Matlab path, you can add them to the path in several ways (see the Paths and Folders section for more information).
Check if your hardware is supported
BrainStream connects to hardware devices via a [FieldTrip buffer]. This connection is controlled by interfacing software. Each acquisition system has its own interfacing software, which is called <system>2ft. For example, the software for the BioSemi acquisition system is called biosemi2ft. See the [FieldTrip website] for a list of hardware systems for which interfacing software is available. If your hardware is supported, you can proceed to set up the connection between BrainStream and your hardware.
Hardware Setup
Interfacing software
Any acquisition system supported by the FieldTrip toolbox can be used as a data source within BrainStream. BrainStream does not connect to the hardware directly, but via a [FieldTrip buffer]. The connection between hardware systems and the FieldTrip buffer is handled by interfacing software. Here, we will show how to use the interfacing software for the BioSemi acquisition system (biosemi2ft). For more information about the interfacing software for other acquisition systems, see the [FieldTrip website].
In BrainStream, the interfacing software is located in the FieldTrip folder: fieldtrip/realtime/acquisition/<system> (you can find the FieldTrip folder by typing bs_folder('FIELDTRIP') in the Matlab command window). Copy this folder to the location where you want to execute biosemi2ft (note that the recorded data will be saved in this location as well).
The BioSemi system needs a configuration (.cfg) file in which some acquisition-related settings are specified. Configuration files for the BioSemi system specify for example from how many channels data should be recorded, and whether the data should be downsampled. Several configuration files for the BioSemi system are available in BrainStream's resources folder (type bs_folder('RESOURCES') in the Matlab command window to find this folder). For more information about configuration files for different acquisition systems, see the [Fieldtrip website].
To start the buffer that connects to the acquisition system, open the terminal (Mac) or command prompt (PC) and go to the folder where you copied biosemi2ft. Then execute:
biosemi2ft <config_file> <gdf-file> <hostname> <port> <config_file>: configuration file <gdf-file>: base name of the gdf file. The suffix .gdf and session counters will be added automatically. <hostname>: optional, default is 'localhost' <port>: optional, default is 1972
When the buffer is connected to the acquisition system, it looks like this:
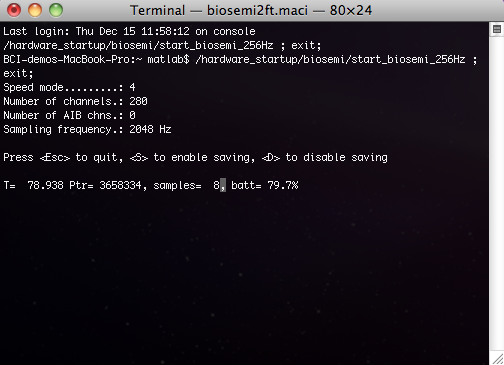
Online Data Manager (ODM)
The biosemi2ft software supports FieldTrip's Online Data Manager, which enables a remote configuration of acquisition settings. For example, you can change the number of streamed or saved channels and filter settings while the biosemi2ft software is running.
In BrainStream, the Online Data Manager allows you to specify acquisition-related settings in your block file. Under topic [<device>], set the OnlineDataManager key to 1 and the remotePort key to the same port value specified at startup of biosemi2ft. You can now specify additional settings as keys under the same topic. For example, under key configureFile, you can specify a configuration file that is different than the one that was used for biosemi2ft startup. BrainStream will send these new configuration settings to the interface software along with a name for the gdf-file saving the raw data.
[biosemi_active2] OnlineDataManager = 1 remotePort = 8000 configureFile = 'biosemi_active2_cap64.cfg'
If the interfacing software <system>2ft does not support the Online Data Manager, the OnlineDataManager key must always be set to 0 (default).
Start and Stop Functions
BrainStream offers the option to specify functions that will be executed either before starting or after stopping streaming data from the hardware device. For example, these functions may start the interfacing software automatically or take care of copying the acquired gdf-file to some dedicated location. You can define the Start and Stop functions in the block file under topic [<device>] and keys startFunction and stopFunction:
[biosemi_active2] startFunction = 'cleanup' stopFunction = 'copy_rawdata'
The functions should take the form:
function my_custom_startFunction(datasource,device) % datasource: reference name of the data source (for example: 'eeg') % device: reference name of the hardware device (for example: 'biosemi_active2') % add code to handle specific tasks, required in your lab before connecting to the fieldtrip buffer function my_custom_stopFunction(datasource,device) % datasource: reference name of the data source (for example: 'eeg') % device: reference name of the hardware device (for example: 'biosemi_active2') % add code to handle specific tasks, required in your lab after completing a block
Hardware markers
Markers can be added to the data either via the hardware or via the FieldTrip buffer (see [.DocsSectionsMarkerHandling Inserting Markers] for more information). A major reason to use hardware instead of buffer markers is the fact that a better synchronization between presented stimuli and data can be achieved. If your hardware device does not support solutions to insert markers via the hardware, you can send all markers via the FieldTrip buffer directly, but note that these markers will be less accurately synchronized with the data.
There are different approaches for inserting hardware markers, depending on the specific hardware device you are using. Markers may for example be inserted via a PC's parallel or serial port or via an input port that is specific to the hardware device. For example, the BioSemi Active Two amplifier has an input trigger channel through which markers can be inserted.
Users should always use BrainStream's [.DocsSectionsProgrmmersGuide#SendHardwareMarker bs_send_hardware_marker] function to send hardware markers to keep code independent of connected hardware. This function will look in the blocksettings for a sndMarker function which actually sends the markers to the hardware device. You must specify this function under topic [<device>] and key SendMarkerFnc. The default sndMarker function is sndBufferMarker, which sends the marker via the FieldTrip buffer (i.e. not via the hardware).
[biosemi_active2] SendMarkerFnc = 'sndBufferMarker' %default
You can write your own sndMarker function for sending markers to your specific hardware device. A custom sndMyMarker function should be in the following format:
function [event, ret] = sndMyMarker(event,m)
% m.name = name of marker
% m.type = type marker
% m.value = marker value (type char!)
% ret > 0 if marker was sent successfully
try
% check m.type and m.value
% ...
% use type and value to sent marker out to the hardware
% send_my_marker(m);
ret = 1;
catch err
ret = 0;
end
An example using a PC's COMM port looks like this:
function [event, ret] = sndCOMMarker(event,m)
mlock % prevent unwanted clearing of persistent variable serobjw
if isempty(serobjw)
serobjw = serial('COM1'); % Creating serial port object
serobjw.Baudrate = 115200; % Set the baud rate at the specific value
set(serobjw, 'Parity', 'none'); % Set parity as none
set(serobjw, 'Databits', 8); % set the number of data bits
set(serobjw, 'StopBits', 1); % set number of stop bits as 1
set(serobjw, 'Terminator', ''); % set the terminator value to newline
set(serobjw, 'OutputBufferSize', 512); % Buffer for write
end
try
fopen(serobjw);
fwrite(serobjw,str2double(m.value)); % write to serial port
fclose(serobjw); % close it
ret = 1;
catch err
disp(err.message);
ret = 0;
end
Note: If your function for sending out markers requires specific resources (both software and hardware), they should also be installed at every connected client. In your experiment design insert a marker from the BS_INIT processing pipeline and define it to execute on the client, which actions then should install required software resources.
Firewall settings
BrainStream uses ports 5001 and 5002 to connect to possible Matlab [.DocsSectionsParallelMode clients]. Furthermore, BrainStream uses ports 1972 and further (up to numbers between 1985-1990) to connect to buffers streaming data (either acquired, simulated, or from file), to install external Matlab modules, or to install buffers related to BrainStream's pre-processing option. Make sure these ports are not blocked by your firewall.