BrainStreamExampleFacesPlaces
Example 3: Decoding Visual Attention with real-time FMRI
[under construction- this is a rough sketch]
We will now describe how to build a real-time fMRI (functional magentic resonance imaging) neurofeedback experiment using Brain Stream. Real-time fMRI experiments are very different compared to real-time EEG experiments. This is because in real-time fMRI, the fMRI scanner cannot be controlled using Brain Stream meaning that data acquistion cannot be started or stopped by Brain Stream and no hardware markers can be sent to the scanner.�
�function event = init_stiminfo(event)
% read [[DataFolder|Data Folder]]
data_folder = bs_get_blockvalue('Experiment','DataFolder');
% add stims folder to Matlab path
addpath(exGenPath(fullfile(data_folder,'stims')));
% tasks should be defined via topic 'Experiment' and key 'Tasks'
for n = 1 : numel(event.tasks)
task = event.tasks{n};
if any(strcmp(task,{'classifier'}))
% doesn't require stimulus definitions
continue
end
% try to load stimulus definitions for this task
try
event.stiminfos.(task) = eval(['get_stiminfo_' task]);
catch err
error('Missing or failed loading definitions for task %s (%s)',task,err.message);
end
end
This function intiliazes the stimulus material for each task. Becasue during the 'Classifier' task (in which the traning data is used to trainn the classsifier,) no stimulus is presented therefore, the code skips loading any stimulus related info during this task.
(from table)
bs_trace('scan_count','add'), bs_trace( 'epoch_count', 'add'), bs_trace( 'sequence_count','add'), bs_trace('alphaLevel','add')
bs_trace.m traces the variable and when these variables change (globally) then it logs the value of the varialbes in the log file. Hence it allows to track what happens to each varaialbe and when.
function event = annotate(event)
% event.stiminfo.num_epochs
% event.stiminfo.num_rest_scans
% event.stiminfo.test.scans_per_epoch
% event.stiminfo.scans_per_epoch;
sc=event.scan_count;
ep=event.epoch_count;
nscans_tsk = event.stiminfo.num_task_scans;
nscans_pre = event.stiminfo.num_pre_scans;
event.scan.task = event.task;
event.scan.data = event.data.raw;
event.scan.sample = event.data.trial.offset;
event.scan.scan_count = event.scan_count;
event.scan.epoch_count = event.epoch_count;
% use booleans for actual task or rest (won't get here during cue)
istask = (sc <= nscans_tsk + nscans_pre);
% start with task, then rest
if istask
% task (training, test, ...)
% only executes when sc > nscans_pre! This is done in the nextScan function.
task_idx = sc-nscans_pre; % scan index during the current actual task
% add label information (add custom labeling via label info field in
% get_stiminfo_<task> function
event.scan.label = event.stiminfo.labels{ep}{task_idx};
else
% rest
event.scan.label = struct('label',0,'info','rest');
end
if strcmp(event.task,'test')
% add labeling for non-training tasks
event.scan.predict = event.classifier.predict; % from bs_test_glmnet
event.scan.X = event.X; % from bs_test_glmnet
event.scan.feedback = event.feedback; % from bs_test_glmnet
end
event.stiminfo contains stimulus information that we make before each task. Annotate function puts this stimulus information for each TR in the structure that is stored once the experiment finish. Hence it makes a data structure that can be used offline to analyze the online real-time run.
function var = putfld(update, var, field) % update: content that should replace content of destination field % dstfield) of global variable % var: global variable % field: field of global variable
Currently dont understand how put work with four input arguments
(G7): put('scan',collect_data,[],'scans')
function event = init_fmri_buffer(event)
numepochs = event.stiminfos.(event.task).num_epochs ;
numscans = event.stiminfos.(event.task).scans_per_epoch;
% initialize fixed cell array buffer to collect scan info structures
event.fmri.scans = init_data(cell(1,numscans*numepochs),[],[],'fixed',2);
end
Make a cell array structure and fills it as scans arrive.
function var = init_data(update, var, field, type, catdim)
% update: size of new buffer
% if update = 'clear', it assumes buffer is already initialized,
% but needs to be filled with all zeros
% catdim: dimension along which to fill the buffer
% type: 'ring', 'fixed', or 'concat'
% Note, for type 'concat', initialize with zero catdim dimension
% to clear the buffer, set update='clear' and specify field if assigned to structure field
if nargin < 3, field = []; end
if nargin < 5, catdim = []; end
% if variable is stored as field of a structure, get it into buf
buf = field2var(var, field);
if ischar(update) && strcmp(update,'clear')
% check type buffer
if isempty(buf) || ~isfield(buf,'isa') || ~strcmp(buf.isa,'buffer_variable')
error('Can not clear an uninitialized buffer');
end
buf.data = zeros(buf.dims);
buf.ptr = 0;
buf.num = 0;
if ~buf.concat
buf.id = ones(1,buf.len)*nan;% fill with nan's to identify empty entries
end
else
buf = buffer_variable(update, type, catdim);
end
% if part of structure, put it back
var = var2field(var,buf,field);
end
Gre-matter mask calculation
The grey matter mask is extracted from the structural scans using SPM8 unified segmentation normailzation procedure. Frist the 192 images of the GRAPPA structural scans are copied from the scanner computer the computer running brainstream. Brain Stream constantly polls the 'dicom' folder and once it detects that all 192 images have been received in the this folder, it starts to segment the structural image into grey-matter, white-matter and CSF. The grey matter mask is high resolution. In order to apply the grey-matter mask to functional scans, it must have the same reoslution and orientation as the functional images. Hence the grey-matter mask is resliced with the first functional scan in the pipeline as reference. After the grey-matter mask is thresholded to 0.5 and then applied to all scans in the tranining and test session.
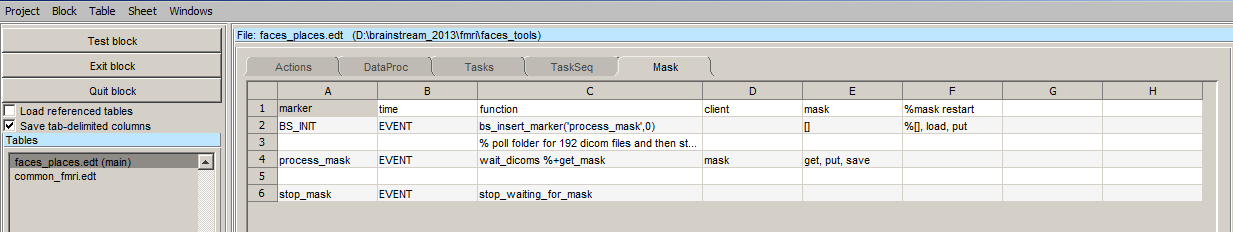
With the start of the experiment using BS_INIT marker, the process_mask marker is inserted. The marker trigger the execution of the wait_dicom function, which polls the dicom folder for 192 dicom images. These dicom images must be transferred to dicom folder via LAN. You can share the dicom folder and make it available on the local area network. Once shared you can then just copy the dicom files from scanner and paste it in the share dicom folder. The rest will be taken care of by brainstream. Once the mask is calculated, the a stop_mask marker is inserted which tells the script to stop pollling for the files.
wait_dicom
function event = wait_dicoms(event)
if isfield(event,'mask') && ~isempty(event.mask)
event = bs_warn(event,'Skipped calculating mask: it is already defined');
return
end
if ~bs_get_blockvalue('Mask','CalculateMask',0)
event = bs_warn(event,'Skipped calculating mask: disabled by user');
return
end
% on error, set mask to empty to let the training part complete
try
uwaittime= 1; % pause 1 seconds between successive polls
% This should contain the path of the folder where the
% DICOMS of the anatomical scans are stored
source_folder = bs_get_blockvalue('Mask','dicom_path');
num_dicoms = bs_get_blockvalue('Mask','NumDicoms');
% poll folder for num_dicoms (192) dicom files and then start the script greyMatterMask
while 1
d = dir(source_folder);
% remove folder entries
d = d(~[d.isdir]);
if ~isempty(d)
% remove entries starting with '.'
entries = {d.name};
N = numel(entries);
d = entries(~cellfun(@strncmp,entries,repmat({'.'},1,N),num2cell(ones(1,N))));
end
if numel(d) < num_dicoms
fprintf('Number of DICOM files: %d, waiting for %d files...\n',numel(d),num_dicoms-numel(d));
% check if BS sends a stop message
if exit_loop
event = bs_warn(event,'Failed calculating mask: stopped waiting for dicom files');
event.mask = [];
return
end
continue
else
break
end
end
% calculate mask
event = get_mask(event);
catch err
showerror(err)
event = bs_warn(event,['Failed calculating mask: ' err.message]);
event.mask = [];
end
function ret = exit_loop()
ret = 0;
% wait for new information in a non-blocking call to the socket
svars = bs_recv_user_brainstream(event,'mask',0,uwaittime);
if ~isempty(svars)
if isempty(svars{1})
ret = 1;
end
end
end
end